Visualisation of Data using R
Author: Darren Teo
About the Author: Year 3 student in NUS in Special Programme in Science
About this tutorial: Just a small little worked example of what one can do to visualise data so people can apply it to their reports / papers / presentations etc. This is by no means an exhaustive list and I am sure there are better ways to go about doing it :))
Section 1: Scatterplots, Boxplots and Grouping by Colour
Scatterplot & Pair plots
Loading in iris dataset
For the purposes of this tutorial, we are going to be using the Iris flower dataset introduced by Ronald Fisher in his 1936 paper The use of multiple measurements in taxonomic problems.
We can try loading in this built-in dataset in R. Like so:
data("iris")
Inspecting the dataset
This dataset contains Sepal and Petal length / width of different Species of flowers Iris setosa, versicolor, and virginica.
We can take a peek at the dataset like this:
head(iris)
| Sepal.Length | Sepal.Width | Petal.Length | Petal.Width | Species | |
|---|---|---|---|---|---|
| 1 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 2 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 3 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 4 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 5 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 6 | 5.4 | 3.9 | 1.7 | 0.4 | setosa |
The next step in exploring datasets is to know and understand what the different variables are referring to, such as Sepal Length, Sepal Width, Petal Lengths and Petal Width!
Optional Information
The dataset numbers are all in centimeters (cm), and the different variables should look foreign to people not well versed in plant morphology. Luckily for us, since this is a well-known dataset, the internet has graphics explaining what they are. 
How handy! This website contains an image on what Sepal / Petal length and widths mean for each row!
Pairs plot: Testing a hypothesis
With this dataset, a hypothesis that we could reasonably come up with is that Sepal.Length and Petal.Width are related in someway. We can visualise this quickly by using the plot() function:
plot(iris$Sepal.Length, iris$Petal.Width)
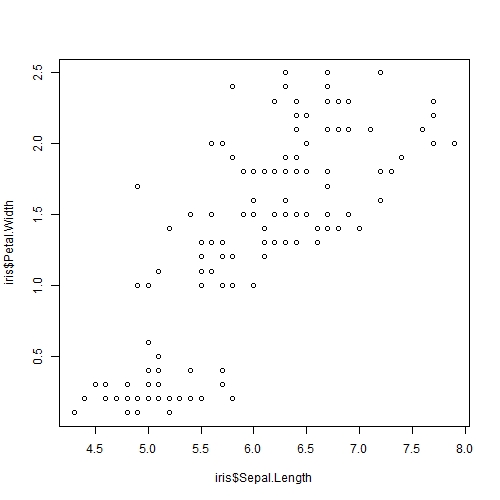
plot()takes in many arguments, but 2 are necessary for a meaningful plot: the x and y values. The arguments must either be provided in that order (x then y, as shown in our example here) or explicitly defined asx = iris$Sepal.Length, y= iris$Petal.Widthin the brackets.- Alternatively,
plot(iris$Petal.Width ~ iris$Sepal.Width)would work as well as it takes the form ofplot(y ~ x)which is a model formula. - Secondly,
plot()also takes an argument oftypewhich tells R which type of plot to be drawn, the default is a scatterplot. However, if a line graph is requiredplot(x,y, type = 'l')would produce a line plot.
Awesome! There seems to be some sort of association. What if we want to explore other combinations of our variables, such as Sepal.Length vs Petal.Length or others? Luckily for us R has a function called pairs() to help us! By adding panel = panel.smooth, R would help us generate a "smooth" curve to help us better visualise correlations!
pairs(iris, panel = panel.smooth)
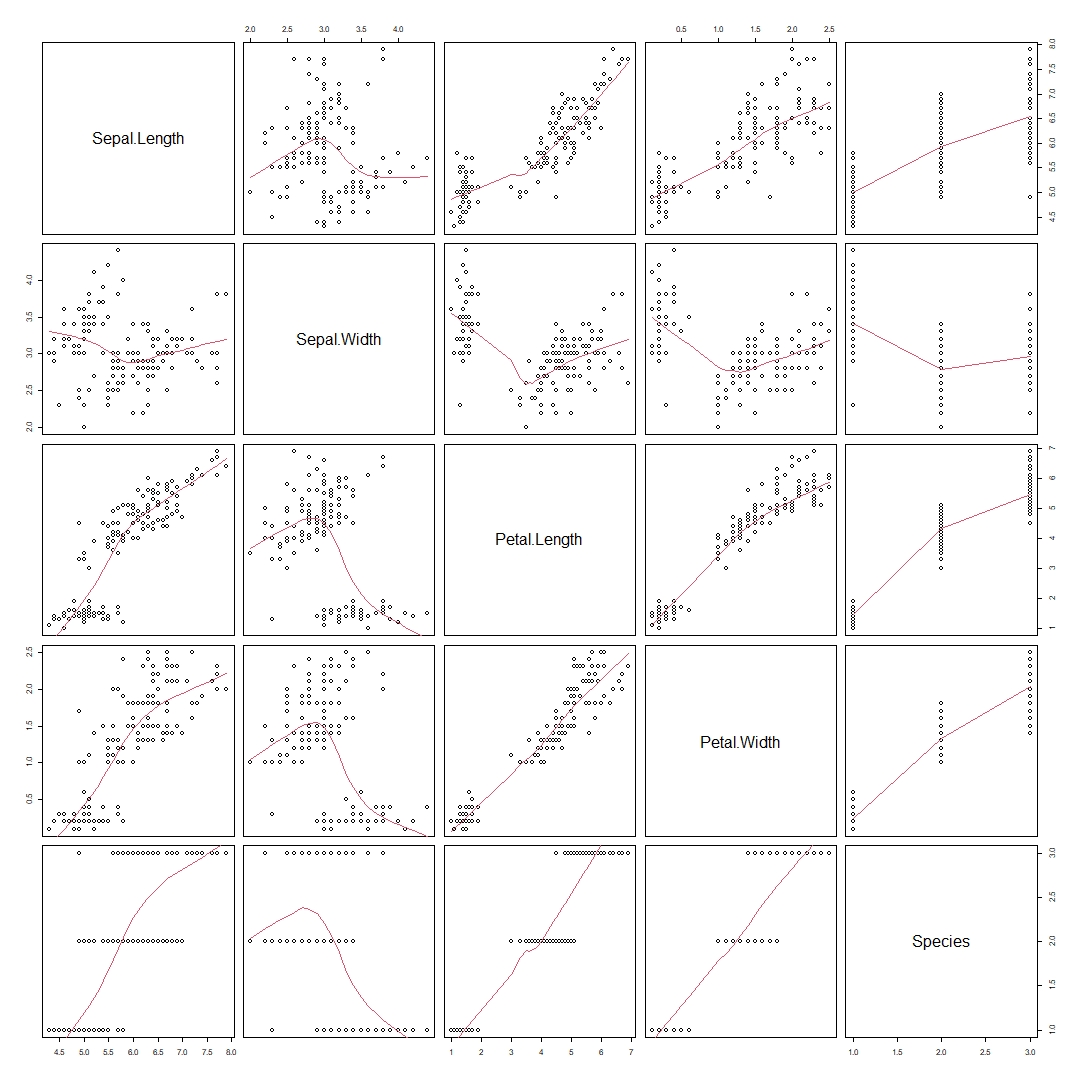
It seems that Petal.Length and Petal.Width has the strongest positive association with each other. This is to be expected as we would expect a flower with a longer petal length to have a longer petal width as well.
It also seems that Sepal.Length and Petal.Length have some sort of correlation as well. Lets use these two variables for our examples later!
Solidifying our research question
Before we proceed any further, it is important in any experiment that we have a solid research question in mind. This is because the type of data presented will help answer your research question.
Lets say we had a basic research question "Are the lengths of the petals associated with the lengths of sepal in I. setosa, I. versicolor, and I. virginica?" So while we're doing data collection, we collected many other variables as well (see table above).
After data collection, we can plot Sepal.Length against Petal.Length. 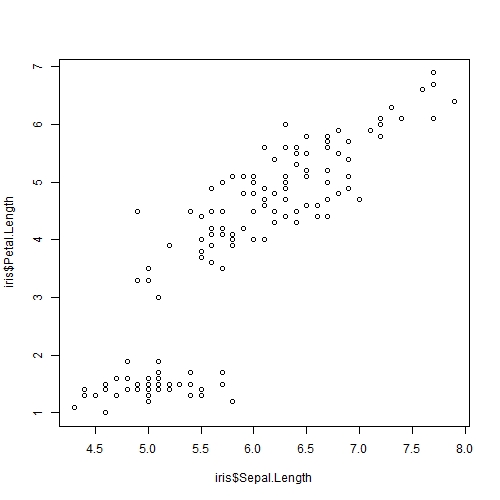
It seems that Petal Length and Sepal Length do have a linear positive correlation with each other!
Scatterplot & Trendlines with ggplot2
If you were Ronald Fisher and wanted to present about the positive correlation of Sepal Length with Petal Length by using the graph presented above, you will probably not succeed in getting your point across.
Luckily, since most of the R community have agreed that base R plotting is terrible for presentations. A very handy package called ggplot2 was created.
If you have not installed ggplot2 in R, you can install it using the following command then load it in.
install.packages("ggplot2") #This is only if you have not installed it before
library(ggplot2)
The cool thing about ggplot2 is that it allows for a very "modular" way of adding to the graph. We first specify the dataframe in which ggplot should look in, in our case iris, followed by the "global" aesthetics layer using aes().
We want our x-axis to be Sepal.Length and y-axis to be Petal.Length
So our base syntax for our example would look like this:
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length))
#To view the graph you can just run
iris_PetalLengthvsSepalLength
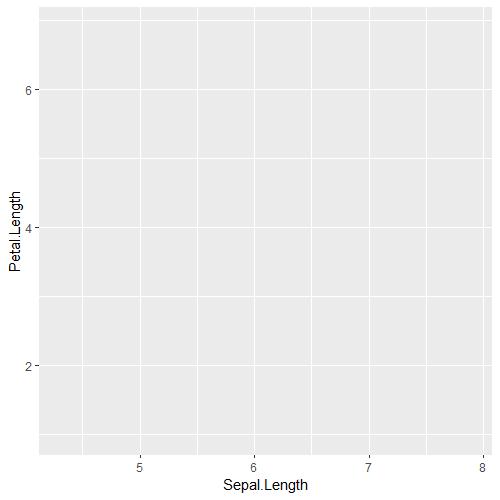
As you can tell, the plot is blank! This is because we only told ggplot to initialise a blank canvas, the next thing we want ggplot to do is to add our points! We can do this by using this geom_point():
#There are two ways to do this. The first way is
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point()
#The second way is, assuming you already defined the variable of iris_PetalLengthvsSepalLength before,
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + geom_point()
#Like always lets view iris_PetalLengthvsSepalLength
iris_PetalLengthvsSepalLength
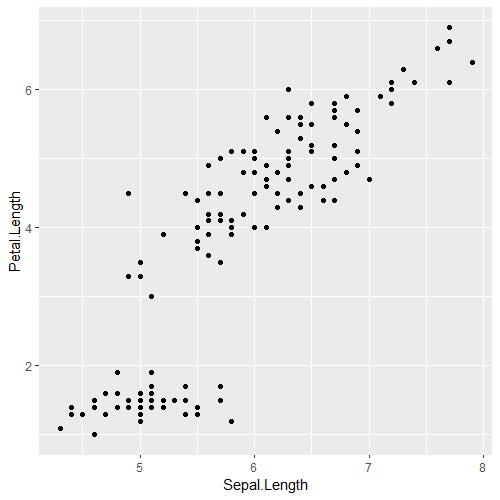
Great, this is the exact same graph as above. Except this has COLOUR. There are a few things we can do to make this graph look more purposeful. For your information, geom_point() can take an additional argument to change the shape and size of your dots if they ever look too small! You can experiment with it yourself, it'll look something like this geom_point(size = 5, shape = 21)
- Label size for the numbers
- Naming of X- and Y-axis
- Title / subtitle
Changing the label size
The first glaring issue is that the labels on the x- and y-axis are very small and may not be easily readable. So let us change that using theme():
#Once again, there are two ways of doing this, you can either build the graph in 1 go
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point() + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25))
#Or make it modular
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + theme() + theme(axis.text=element_text(size=20))
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + theme(axis.title=element_text(size=25))
#Like always lets view iris_PetalLengthvsSepalLength
iris_PetalLengthvsSepalLength
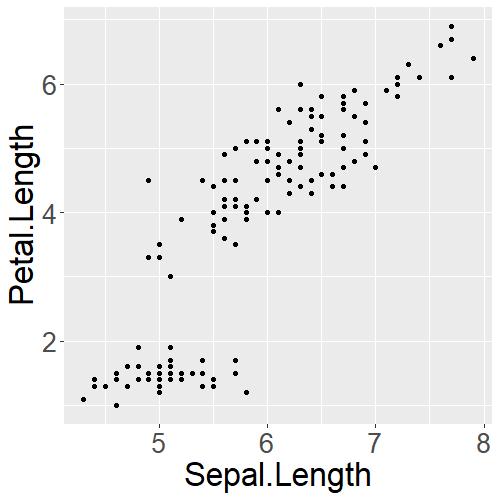
theme(axis.text=element_text(size=20)) changes the numbers on the x- and y-axis whereas theme(axis.title=element_text(size=25)) changes the size of the x- and y-axis labels!
- Label size for the numbers
- Naming of X- and Y-axis
- Title / subtitle
Naming of X- and Y-axis & Title / subtitle
Luckily for us, since the dataframe is named appropriately, most readers would know the x- and y-axis are representing Sepal and Petal length. BUT, readers would not know the units. It is often good practice to rename your axis to be readable by everyone.
Lets rename the axis so that they portray the right information from the get go.
#You can either do this
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point() + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)')
#or do this if you already have iris_PetalLengthvsSepalLength defined earlier.
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)')
#Like always lets view iris_PetalLengthvsSepalLength
iris_PetalLengthvsSepalLength
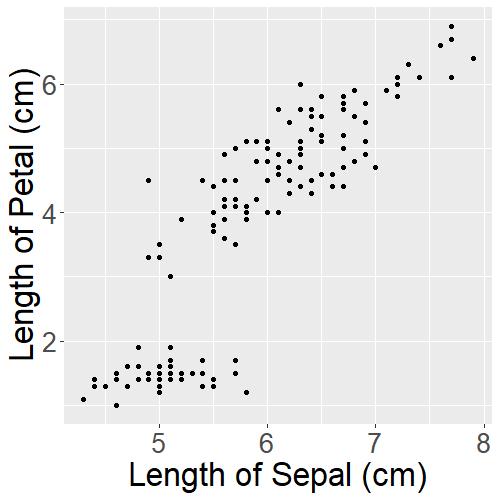
Now with one look, readers can get an idea of what the graph is about. But to make it even clearer, we will need to add a title to the graph:
#Because Iris is the genus, when typing it, they need to be italicised this code snippet below will give you an example.
subtitle_iris = expression(paste("of various ",italic("Iris "), 'flower species'))
#Once again, you can do this
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point() + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)') +
labs(title = "Length of Petals (cm) vs Length of Sepals (cm)", subtitle = subtitle_iris) +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20))
#Or do this to add on
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + labs(title = "Length of Petals (cm) vs Length of Sepals (cm)", subtitle = subtitle_iris)
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + theme(plot.title = element_text(size=30))
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + theme(plot.subtitle = element_text(size=20))
#Like always lets view iris_PetalLengthvsSepalLength
iris_PetalLengthvsSepalLength
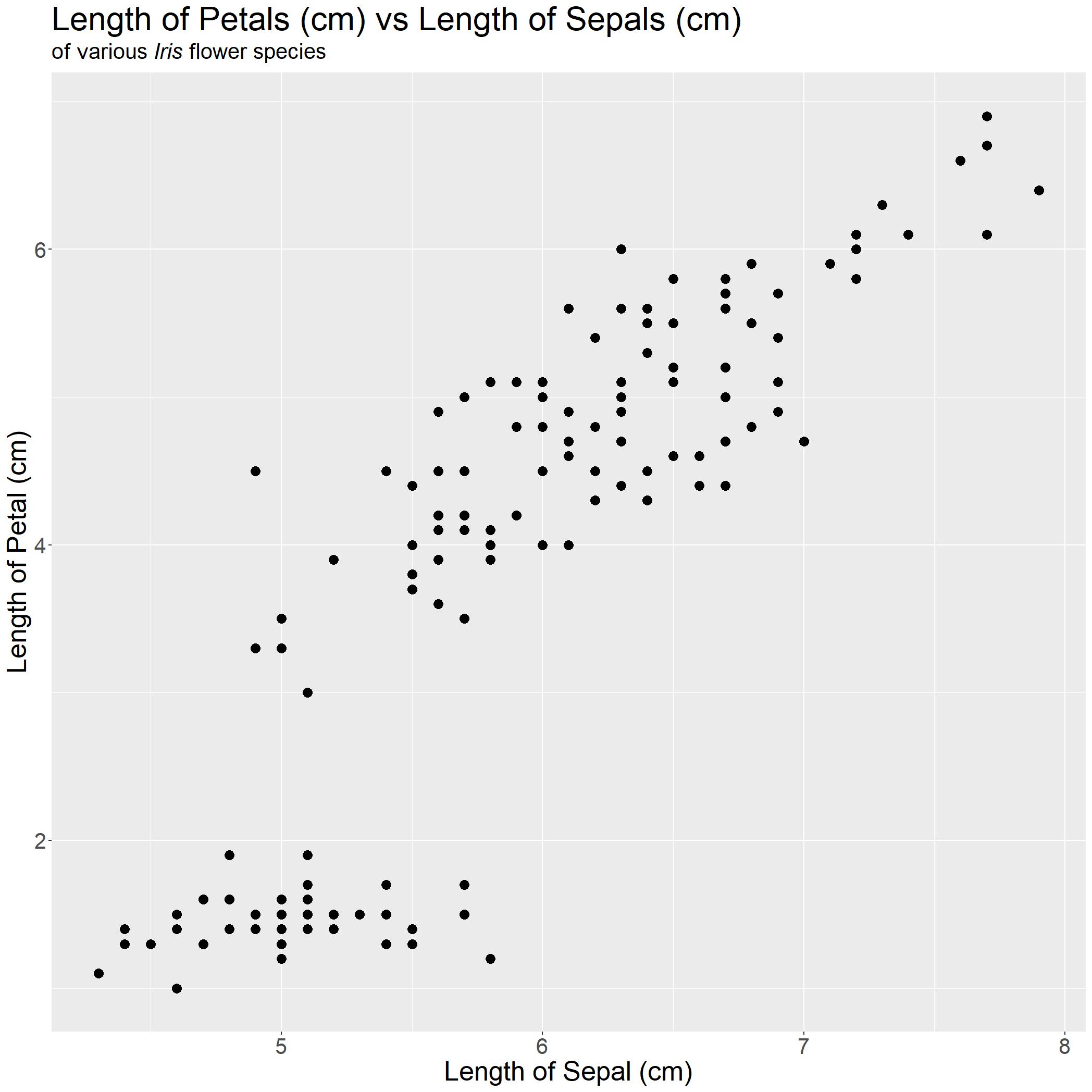
Editor note: I had to increase the size of the graph here while saving so I changed the size of the points by using the command
geom_point(size = 4)
Cool, our checklist is done!
- Label size for the numbers
- Naming of X- and Y-axis
- Title / subtitle
Giving meaning to the graph
We have a decent graph generated, but it does not really tell the reader anything immediately. Linking back to our research question, we want to show that there is a linear association between length of petal and length of sepal of various Iris flower species. The easiest way we can do this is by adding a best-fit linear regression line!
#Once again, you can do this
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point() + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)') +
labs(title = "Length of Petals (cm) vs Length of Sepals (cm)", subtitle = subtitle_iris) +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20)) +
stat_smooth(method='lm')
#Or do this to add on
iris_PetalLengthvsSepalLength = iris_PetalLengthvsSepalLength + stat_smooth(method='lm')
#Like always lets view iris_PetalLengthvsSepalLength
iris_PetalLengthvsSepalLength
#The dark grey borders around the blue linear line indicates the confidence interval for each point on that line.
#If you would like to remove, you can use this function call
#stat_smooth(method='lm', se = FALSE) instead of stat_smooth(method='lm')
#the method = 'lm' here specifies which model to fit the data with!
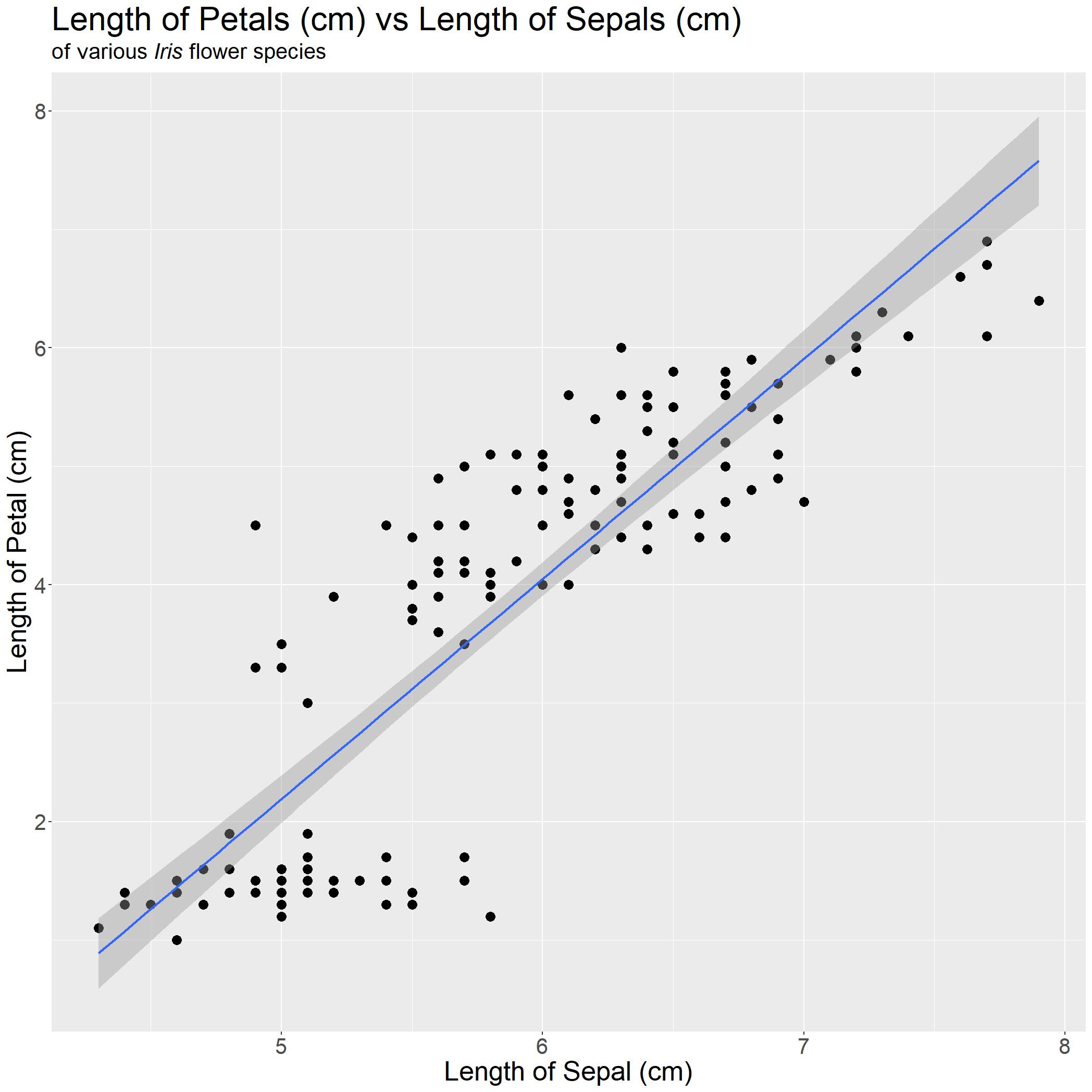
Let us compare with our base graph from earlier to the one we have now!
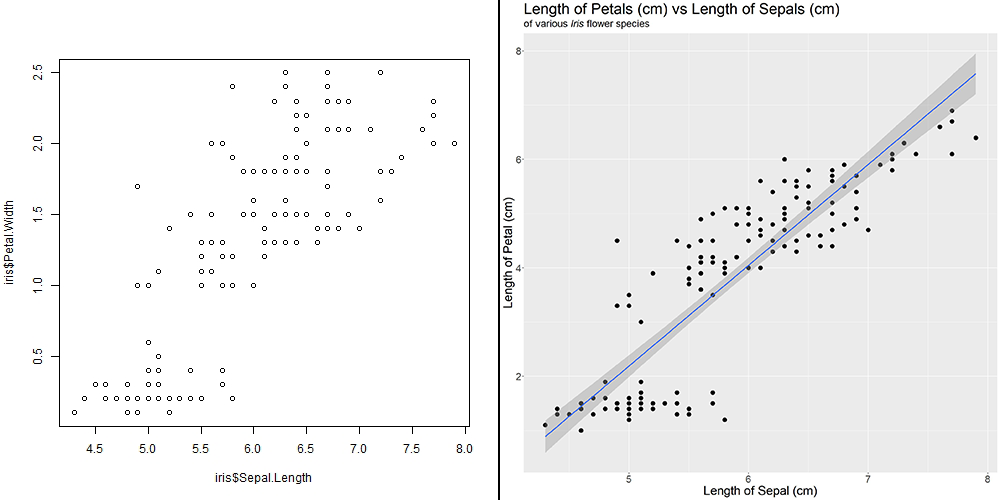
With one look, readers can tell exactly what they are looking at and what message you would want them to takeaway! In this case, as Sepal Length increases, Petal Length increases. This is important, as the use of graphs (and graphics) can greatly assist the audience in learning or understanding the story you want them to take away.
Additional info
In the event that you have other ways you would like to fit your data (since not all data are linearly associated), you can still fit your data after generating a model. In this example, I have generated a linear regression model, but it can be any type of model and it'll still work!
#Generating the simplest linear regression model
iris_lm = lm(Petal.Length ~ Sepal.Length, data = iris)
summary(iris_lm)
#Generating datapoints and putting them in the same dataframe as iris for accessibility
iris.predict = cbind(iris, predict(iris_lm, interval = 'confidence'))
iris_withlm = ggplot(iris.predict, aes(x=Sepal.Length, y = Petal.Length))+
geom_point(size = 4) +
geom_line(aes(Sepal.Length, fit),color="blue", size = 1.5) + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)') +
labs(title = "Length of Petals (cm) vs Length of Sepals (cm)", subtitle = subtitle_iris) +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20))
iris_withlm
Boxplots
Visualising Categorical Variables
In the previous section, both variables shown are continuous variables, which means they can be any continuous value, like numbers. What if one of the variables is categorical, like Species?
In the pairs() plot that is provided all the way to the top, you would notice that the Species of Iris has some form of correlation with both Sepal.Length and Petal.Length.
So, if our research question was to investigate the Sepal.Length and Petal.Length (continuous variable) between Species (categorical variables), we simply need to modify some of our code from the previous section to work!
One of the ways to visualise continous variables against categorical variables is to use a boxplot.
For the sake of simplicity of the tutorial, I would not be adding theme().
#This was the previous code showing continous variable against another continuous variable
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point() +
stat_smooth(method='lm')
iris_PetalvsSpecies = ggplot(iris, aes(x = Species, y = Petal.Length)) +
geom_boxplot()
iris_SepalvsSpecies = ggplot(iris, aes(x = Species, y = Sepal.Length)) +
geom_boxplot()
#Call these two graphs to view them... right?
iris_PetalvsSpecies
iris_SepalvsSpecies
Plotting 2 ggplot graphs in the same window
Bummer. We cannot visualise these two graphs at the same time! Your first instinct might be to use par(mfrow=c(1,2)) but that will not work for ggplot2 graphs. What we need is gridExtra
Install gridExtra and load it if you have not:
install.packages('gridExtra')
library(gridExtra)
Then displaying the graphs is as simple as 1 line of code:
grid.arrange(iris_PetalvsSpecies,iris_SepalvsSpecies,ncol=2)
#There are fancier ways of arranging your graphs using grid.arrange() using matrices
#But I will not be covering it here as it will take very long
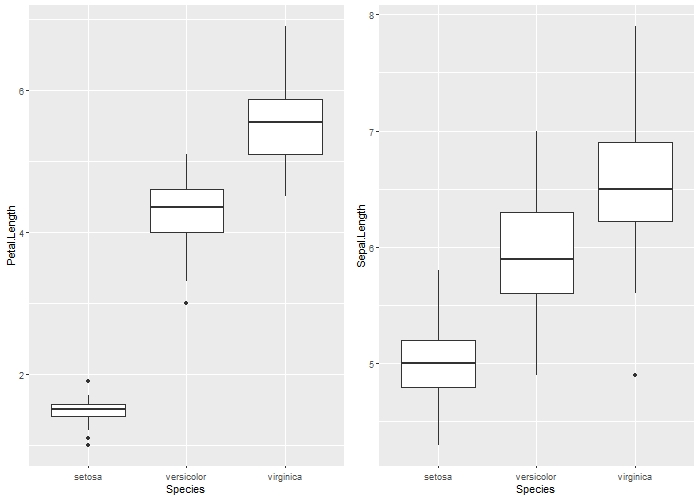
It is clear that there is some form of association between both Sepal.Length and Petal.Length and Species, with I. virginicca having the longest Petal.Length and Sepal.Length and I. Setosa having the shortest.
However, the actual numbers are unknown and ONLY the general trend can be seen!
So, how do we combine all these information together into a single graph?
Scatterplot: Grouping by Colour
From two sections ago, we could see, on average across all species of Iris flowers, Sepal.Length is positively associated with Petal.Length. However, we have to be careful with this conclusion as it is prone to ecological fallacy. As we are making conclusions on a group and we might conclude this same trend within each species of Iris flowers.
So, you might be asking, how do we visualise this when we only have x- and y-axis on a graph? The secret lies in the COLOR or SHAPE of each data point!
Let me show you what I mean, remember our previous code?
iris_PetalLengthvsSepalLength = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length)) +
geom_point() + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)') +
labs(title = "Length of Petals (cm) vs Length of Sepals (cm)", subtitle = subtitle_iris) +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20)) +
stat_smooth(method='lm', se = FALSE)
We can add an additional argument in the global aes() layer to group all points by species by other COLOR or SHAPE like so:
#Grouping by colour
iris_PLSL_col = ggplot(iris, aes(x=Sepal.Length, y = Petal.Length, col = Species)) +
geom_point() + theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Length of Sepal (cm)') + ylab('Length of Petal (cm)') +
labs(title = "Length of Petals (cm) vs Length of Sepals (cm)", subtitle = subtitle_iris) +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20)) +
stat_smooth(method='lm', se = FALSE)
#If you would like to group by shape, the argument col can be changed from col to shape.
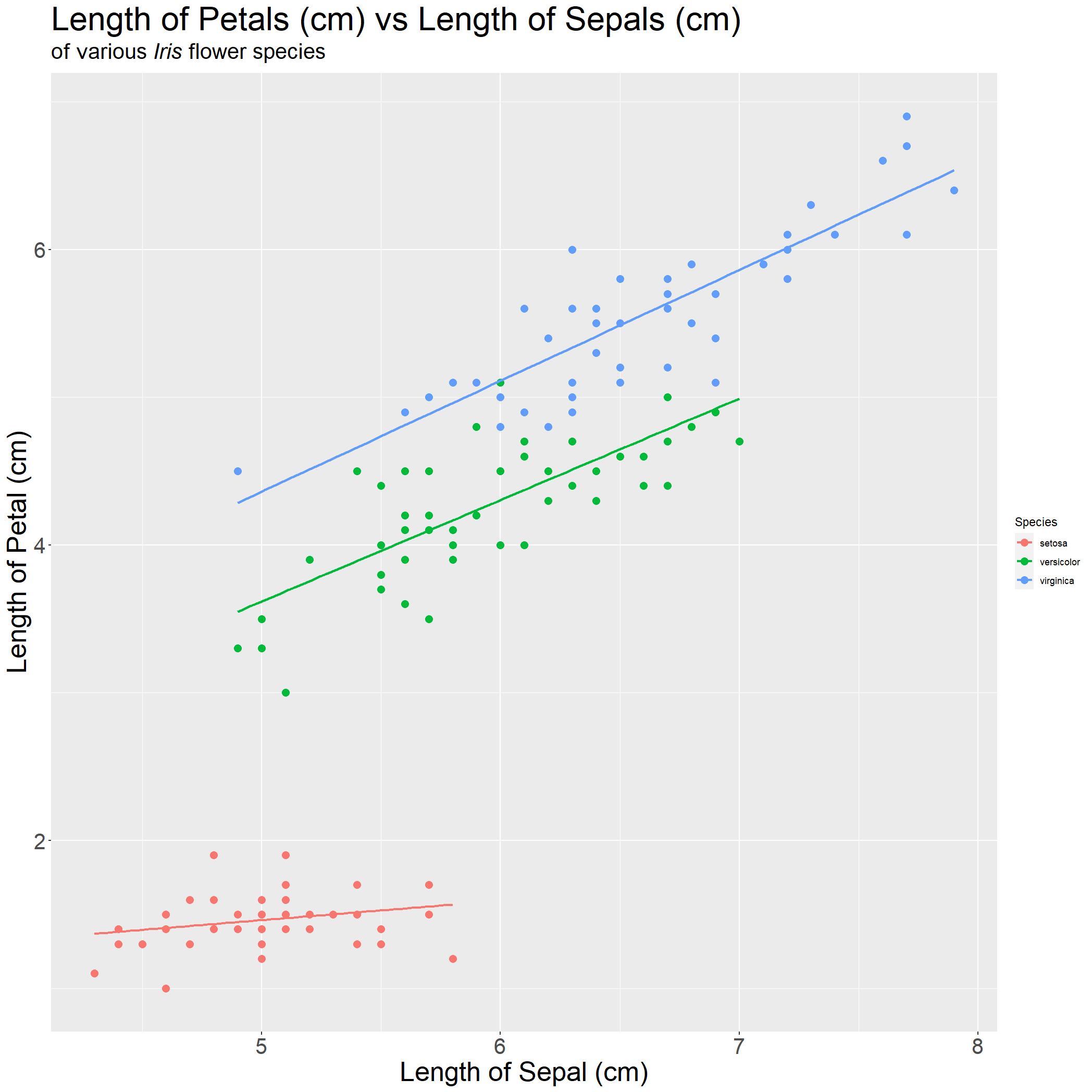
It seems like all three species of Iris are positively associated. With red being I. Setosa, green being I. Versicolor and blue being I. virginica. There is one thing left! Thats right, the legend size. Lets fix that real quick
iris_PLSL_col = iris_PLSL_col + theme(legend.text = element_text(size = 20)) + theme( legend.title = element_text(size = 25))
#There are other aspects you can tweak for the legend, such as background colour and location!
iris_PLSL_col
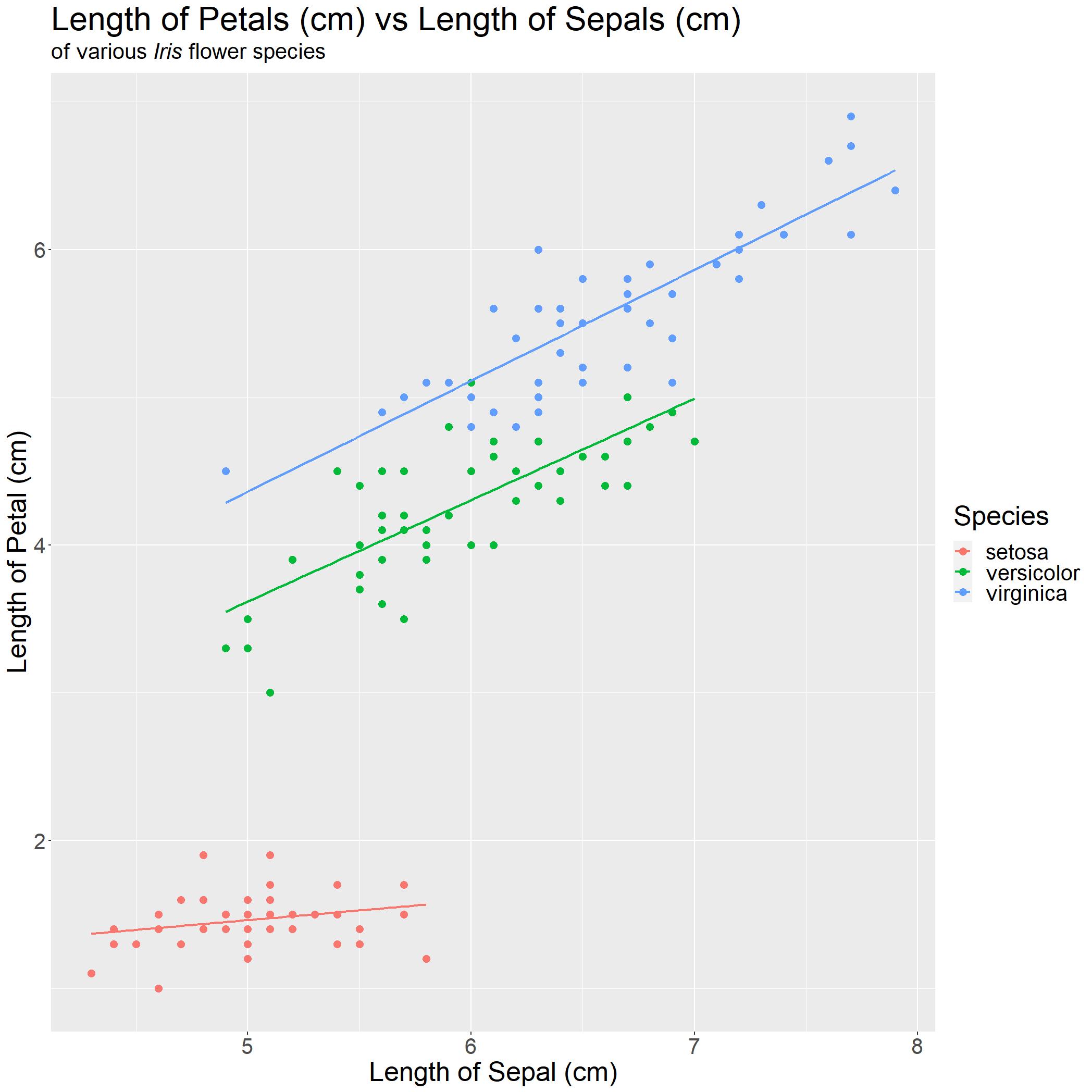
We are done! With a quick glance, anyone can immediately understand the following points:
- Each species of Iris flower, the
Petal LengthandSepal Lengthare positively associated - Weak association for I. setosa
- Strong association for I. versicolor and I. virginica
- I. virginica have the longest Petal and Sepal on average as compared to the other three species.
Saving graphs
To save your ggplot2 graphs, you can use the following command(s):
#Where this image will be saved can be found in the directory after running getwd()
getwd()
#alternatively, you can set the working directory using setwd()
setwd('C:\\My\\New\\Directory')
iris_PLSL_col
ggsave('FILE_NAME.jpg', width = 2096, height = 2096, units = 'px', dpi = 300)
#more detailed options can be seen after running ?ggsave
If however, you are using the grid.arrange() function to generate your graph, you would need to utilise the following commands:
jpeg("FILE_NAME.jpg",quality = 100,width = 1028, height = 1028, units = "px")
grid.arrange(ggplot_graph1,ggplot_graph2,ncol=2)
dev.off()
Addtional information
Do note, that there can be TOO MUCH information on a single graph. Generally, representing the data on x- and y- axis and grouping the points accordingly by colour is the maximum I would go for any graph.
If I would want to explore another variable that I have collected, I would generate another graph at that point.
Bad examples of graphs
To illustrate this point, I added another grouping factor of Sepal.Width for colour and shifted Species to shape.
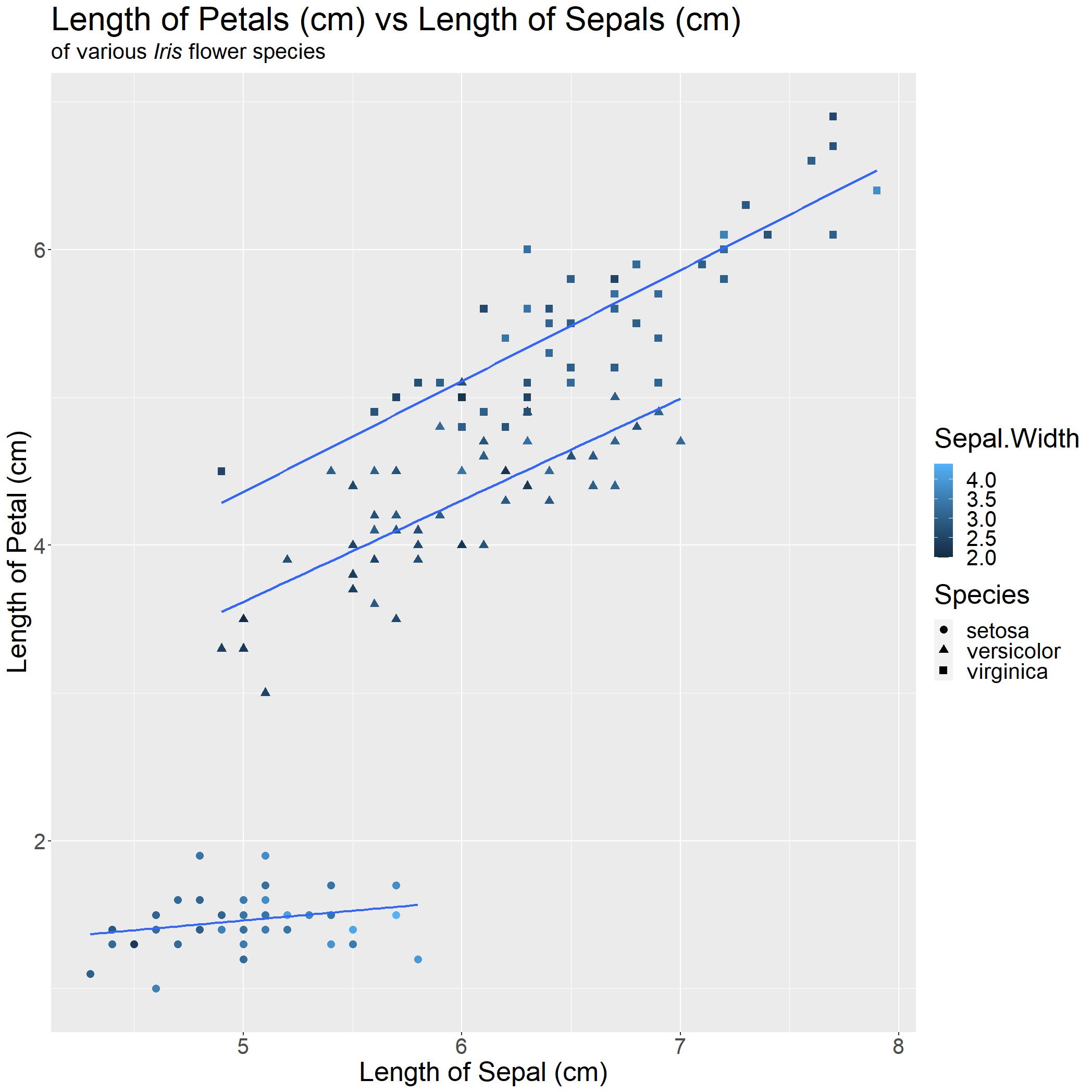
With only a quick glance, you cannot tell much. This example serves to show that "more isn't always better" and that the way the data is presented would aid in the readability of your point.
Now, this is a really exagerrated example of what NOT to do…
Theres just too much text / useless information which could have been in a table and it distracts the reader from the main point of the graph, notwithstanding the overlapping of text affecting readability.
Section 2: Histograms, Barplots and Grouping by Types
In this section, I am going to attempt to visualise data from the birthwt dataset, which was collected at Baystate Medical Center, Springfield, Mass in 1986.
Histograms
Loading in birthwt dataset from MASS
This particular dataset can be found in MASS package and we can load it in like so
library(MASS)
head(birthwt)
| low | age | lwt | race | smoke | ptl | ht | ui | ftv | bwt | |
|---|---|---|---|---|---|---|---|---|---|---|
| 85 | 0 | 19 | 182 | 2 | 0 | 0 | 0 | 1 | 0 | 2523 |
| 86 | 0 | 33 | 155 | 3 | 0 | 0 | 0 | 0 | 3 | 2551 |
| 87 | 0 | 20 | 105 | 1 | 1 | 0 | 0 | 0 | 1 | 2557 |
| 88 | 0 | 21 | 108 | 1 | 1 | 0 | 0 | 1 | 2 | 2594 |
| 89 | 0 | 18 | 107 | 1 | 1 | 0 | 0 | 1 | 0 | 2600 |
| 91 | 0 | 21 | 124 | 3 | 0 | 0 | 0 | 0 | 0 | 2622 |
Optional Information
According to the R documentation, these are what the variables represent:
lowindicator of birth weight less than 2.5 kg.agemother's age in years.lwtmother's weight in pounds at last menstrual period.racemother's race (1 = white, 2 = black, 3 = other).smokesmoking status during pregnancy (0 = not smoking, 1 = smoking).ptlnumber of previous premature labours.hthistory of hypertension.uipresence of uterine irritability.ftvnumber of physician visits during the first trimester.bwtbirth weight in grams.
Solidifying our research question
Just like in the previous section, it is important in any experiment that we have a solid question in mind. For the sake of showcasing histograms and grouped histograms, we can have a basic research question of "Are the birth weight (g) bwt of babies affected by the smoking status of mothers during pregnancy smoke?"
Facet_grid: Grouped Histograms
Hence, our two desired variables are bwt and smoke. Since we have already gone through boxplots showing general trends, we are going to make use of grouped histograms to show more numbers.
This is done by utilising the facet_grid() function in ggplot
birthwt_hist = ggplot(birthwt, aes(x=bwt)) +
geom_histogram(fill="white", colour="black") +
facet_grid(smoke ~ .) +
#This is just for making the graph look nice!
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Baby Birth Weight (g)') + ylab('Frequency') +
labs(title = "Frequency of Baby Birth Weight (g) vs Smokers and Non-smokers") +
theme(plot.title = element_text(size=30)) +
#This is just for changing the text size for grouping
theme(strip.text = element_text(size = 20))
birthwt_hist
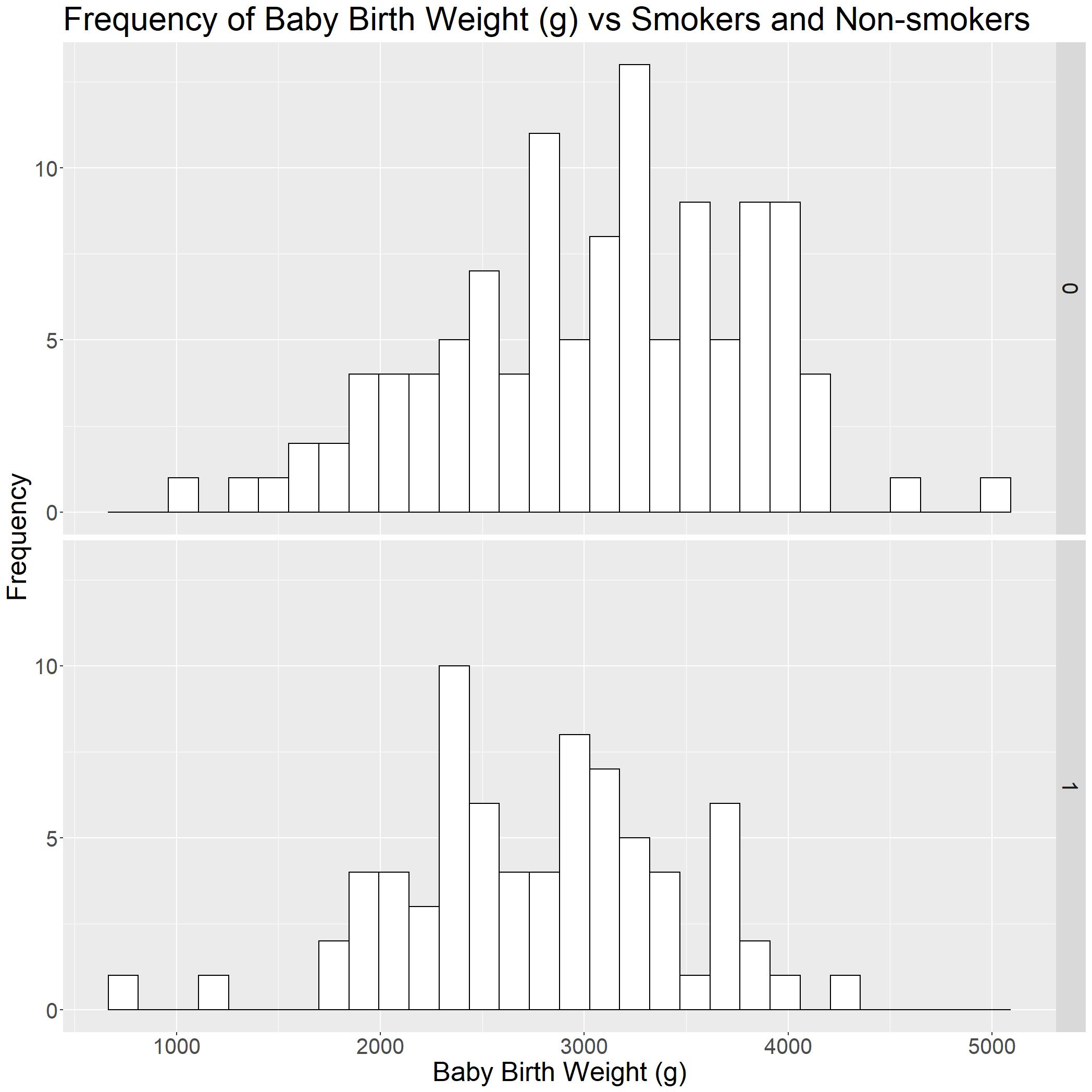
Now that we have a basic histogram, we can roughly tell that the baby birth weight (g) (bwt) is higher when mothers are not smoking (smoke = 0).
However, while presenting data, the levels of 0 and 1 may not be highly intuitive for readers. Hence, we would need to change it.
We would first need to add a new column SmokeR to help us rename 0 and 1 using the factor() function. The first argument takes in the column which R wants to use, the second argument of c('0','1') takes in the levels you want to change and lastly, the third argument takes in a vector to which to rename 0 and 1 in that order.
birthwt$smokeR = factor(birthwt$smoke, c('0','1'), c('No Smoke', 'Smoke'))
head(birthwt)
| low | age | lwt | race | smoke | ptl | ht | ui | ftv | bwt | smokeR | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 85 | 0 | 19 | 182 | 2 | 0 | 0 | 0 | 1 | 0 | 2523 | No Smoke |
| 86 | 0 | 33 | 155 | 3 | 0 | 0 | 0 | 0 | 3 | 2551 | No Smoke |
| 87 | 0 | 20 | 105 | 1 | 1 | 0 | 0 | 0 | 1 | 2557 | Smoke |
| 88 | 0 | 21 | 108 | 1 | 1 | 0 | 0 | 1 | 2 | 2594 | Smoke |
| 89 | 0 | 18 | 107 | 1 | 1 | 0 | 0 | 1 | 0 | 2600 | Smoke |
| 91 | 0 | 21 | 124 | 3 | 0 | 0 | 0 | 0 | 0 | 2622 | No Smoke |
birthwt_histR = ggplot(birthwt, aes(x=bwt)) +
geom_histogram(fill="white", colour="black") +
facet_grid(smokeR ~ .)+
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Baby Birth Weight (g)') + ylab('Frequency') +
labs(title = "Frequency of Baby Birth Weight (g) vs Smokers and Non-smokers") +
theme(plot.title = element_text(size=30)) +
theme(strip.text = element_text(size = 20))
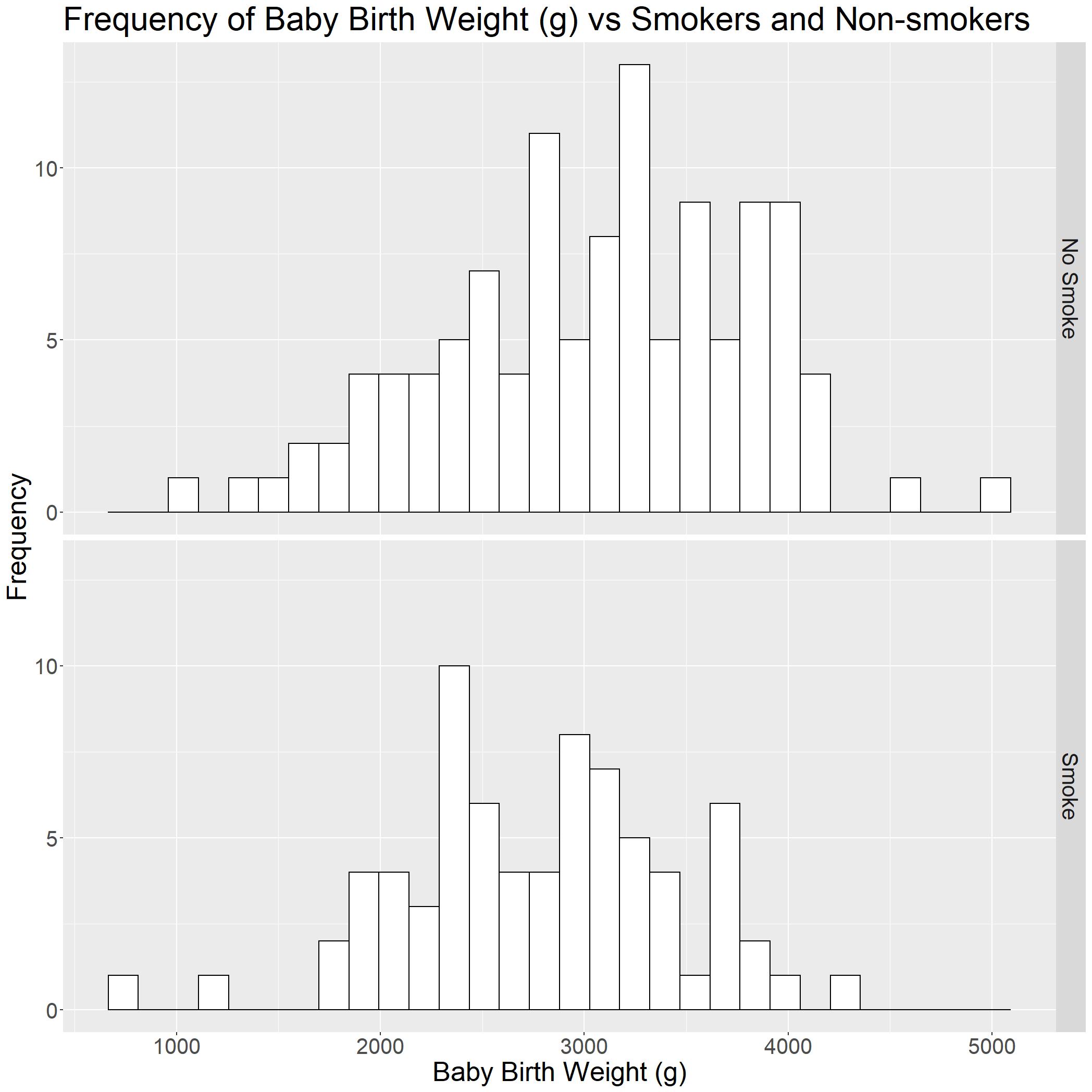
This is much clearer now!
Histograms: grouping with colours
What if the comparison of using a grouped histogram is not that obvious? It might be better to put them all in the same axis and use colour to separate the two. In this case, we would need to make some adjustments.
The important thing to take not of here is while calling the geom_histogram() function. We have the position = 'dodge' here so that histogram are not overlapping with each other.
Leaving position argument out would automatically make the histograms stack ontop of each other. There are other arguments for histograms such as identity.
birthwt_histc = ggplot(birthwt, aes(x=bwt, fill=smokeR)) +
geom_histogram(position="dodge", alpha=0.4)+
#Making the graph look nice.
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Baby Birth Weight (g)') + ylab('Frequency') +
labs(title = "Frequency of Baby Birth Weight (g) vs Smokers and Non-smokers") +
theme(plot.title = element_text(size=30)) +
#Changing the aesthetics of the legend
theme(legend.text = element_text(size = 20)) +
theme(legend.title = element_text(size = 25))+
#Changing the legend title from 'SmokeR' to 'Smoking Status'
labs(fill='Smoking Status') +
#Specifying the position of the legend to the top right
theme(legend.position = c(0.9, 0.9))+
#Removing the legend background from the original white rectangle.
theme(legend.background=element_blank())
birthwt_histc
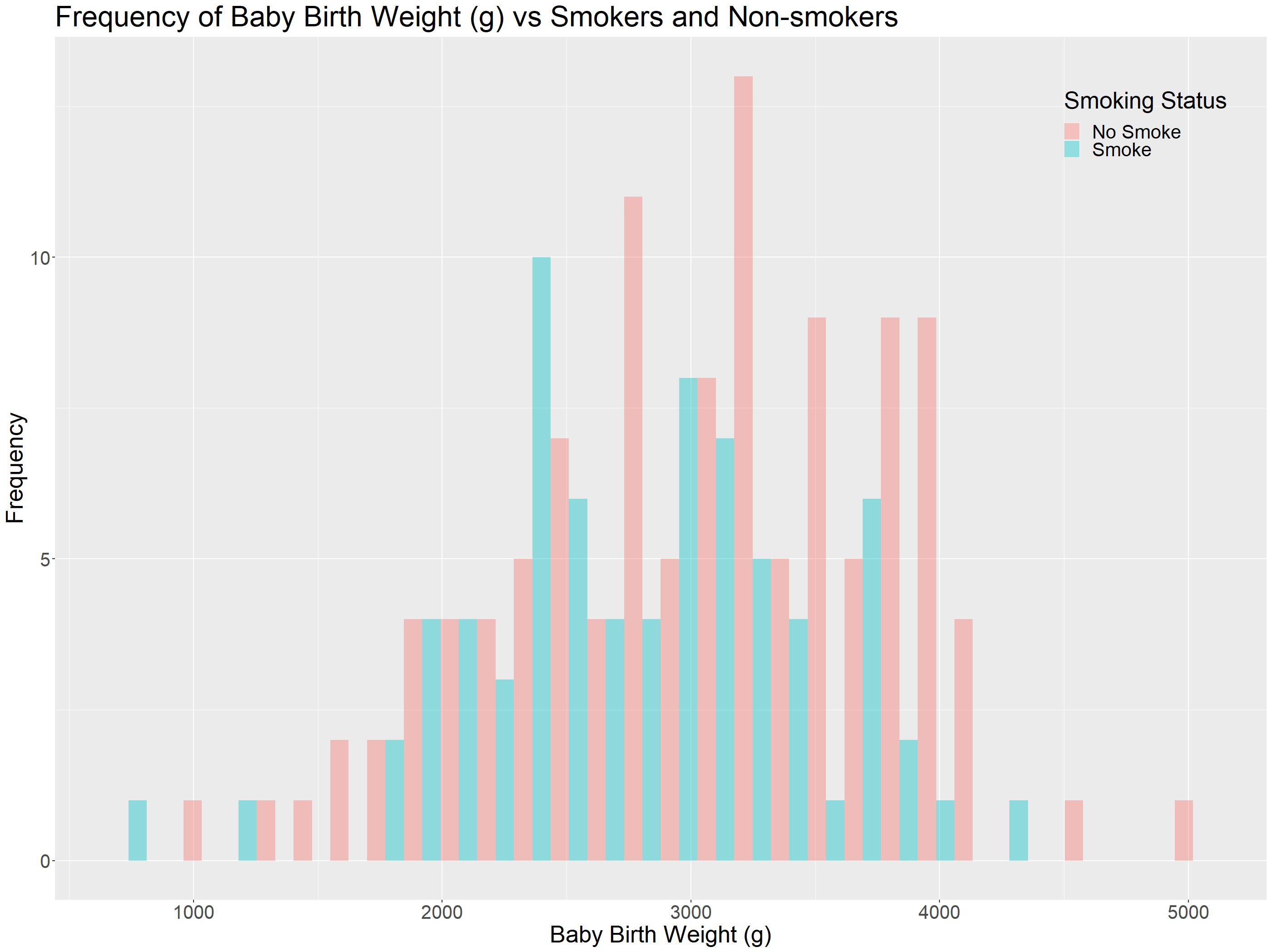
Histogram Presentations: Colour Selection / Focusing
In presentations, sometimes you would need to highlight different groups in this histogram, we are going to look at the function scale_fill_manual() to do so!
The function scale_fill_manual() accepts many argument, one of them is values. In the code snippet below, we set No Smoke to the familiar red using HTML color code, and set Smoke to grey:
birthwt_histc1 = ggplot(birthwt, aes(x=bwt, fill=smokeR)) +
geom_histogram(position="dodge", alpha=0.4)+
scale_fill_manual(values=c("#F8766D", "grey"))+
#Making the graph look nice.
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Baby Birth Weight (g)') + ylab('Frequency') +
labs(title = "Frequency of Baby Birth Weight (g) vs Smokers and Non-smokers") +
theme(plot.title = element_text(size=30)) +
#Changing the aesthetics of the legend
theme(legend.text = element_text(size = 20)) +
theme(legend.title = element_text(size = 25))+
#Changing the legend title from 'SmokeR' to 'Smoking Status'
labs(fill='Smoking Status') +
#Specifying the position of the legend to the top right
theme(legend.position = c(0.9, 0.9))+
#Removing the legend background from the original white rectangle.
theme(legend.background=element_blank())
birthwt_histc1
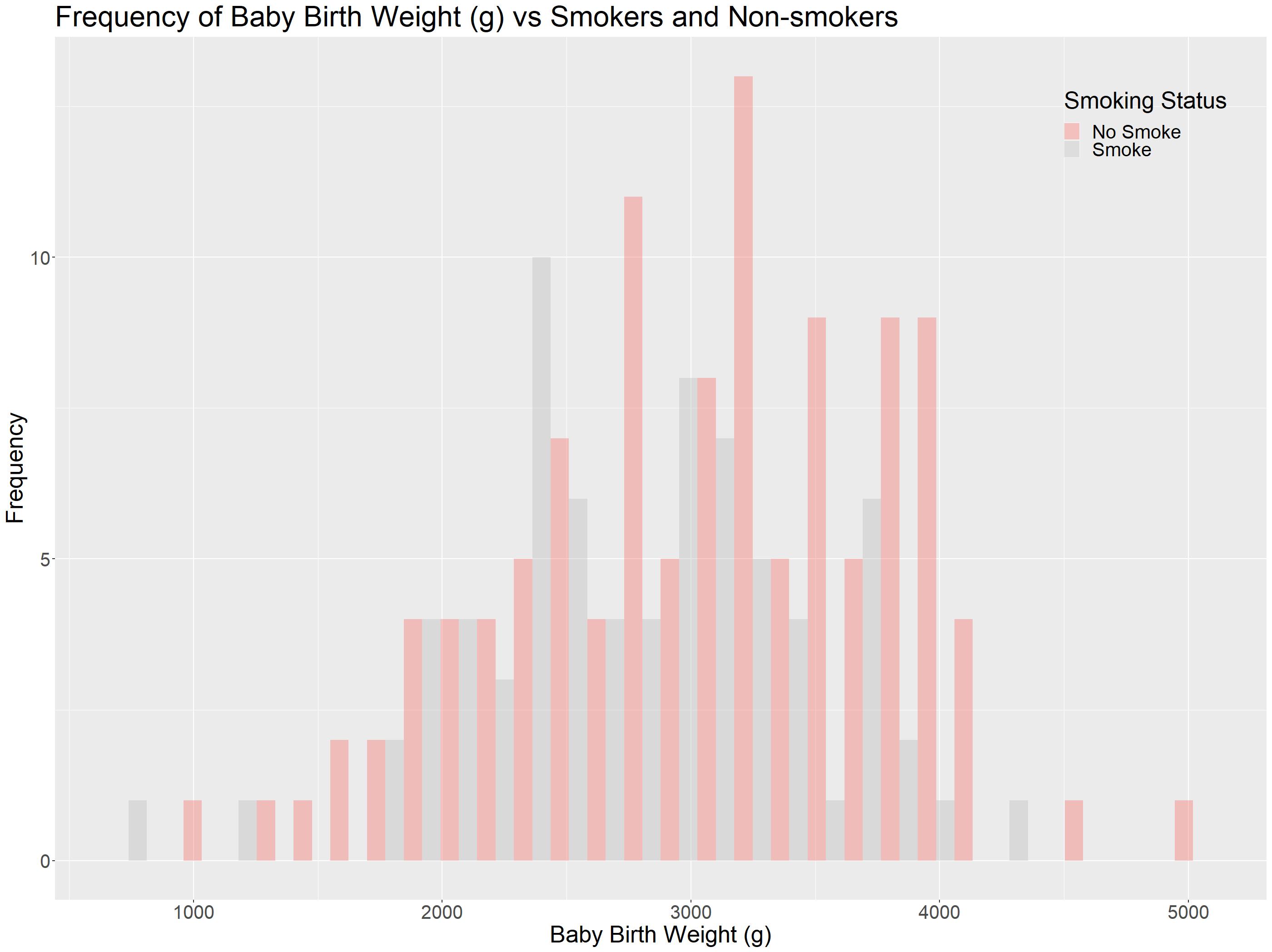
This way, we can highlight just the Non-Smokers!
Likewise, if we want to highlight the Smokers, we can change the scale_fill_manual() to scale_fill_manual(values=c("#grey", "#00BFC4")):
birthwt_histc2 = ggplot(birthwt, aes(x=bwt, fill=smokeR)) +
geom_histogram(position="dodge", alpha=0.4)+
scale_fill_manual(values=c("#grey", "#00BFC4"))+
#Making the graph look nice.
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Baby Birth Weight (g)') + ylab('Frequency') +
labs(title = "Frequency of Baby Birth Weight (g) vs Smokers and Non-smokers") +
theme(plot.title = element_text(size=30)) +
#Changing the aesthetics of the legend
theme(legend.text = element_text(size = 20)) +
theme(legend.title = element_text(size = 25))+
#Changing the legend title from 'SmokeR' to 'Smoking Status'
labs(fill='Smoking Status') +
#Specifying the position of the legend to the top right
theme(legend.position = c(0.9, 0.9))+
#Removing the legend background from the original white rectangle.
theme(legend.background=element_blank())
birthwt_histc2
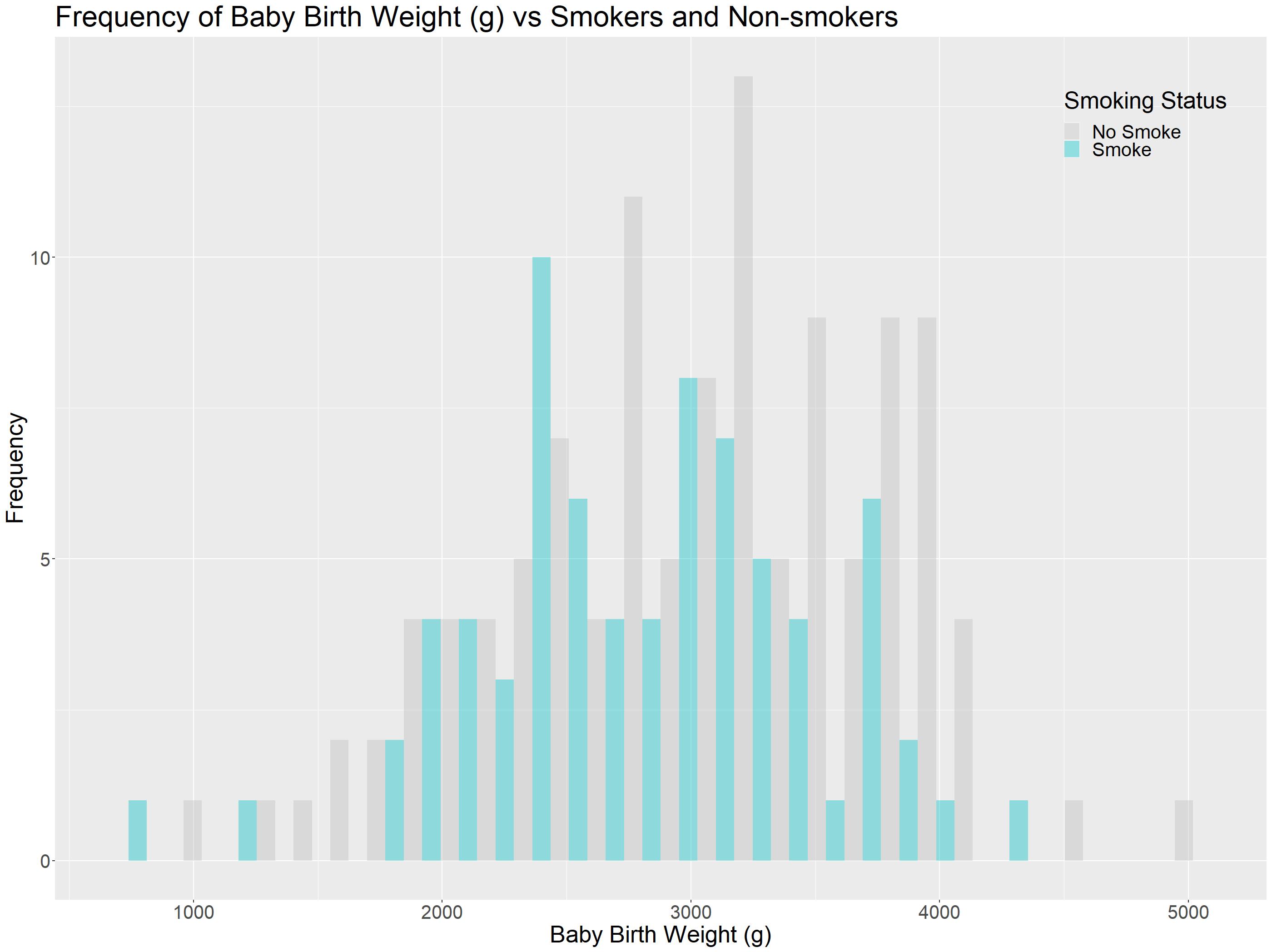
Grouped Bar Charts
Similarly to Section 1, there is a chance that we fall into an ECOLOGICAL FALLACY. Hence, we would need to group some of our data together. In our tutorial, we are going to group them by race of mothers.
Just like our smoke variable, the race of the mother are also in 1, 2 and 3. We would need to refactor that using factor() like before!
birthwt$raceF = factor(birthwt$race, c('1','2','3'), c('White', 'Black', 'Other'))
head(birthwt)
| low | age | lwt | race | smoke | ptl | ht | ui | ftv | bwt | smokeR | raceF | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 85 | 0 | 19 | 182 | 2 | 0 | 0 | 0 | 1 | 0 | 2523 | No Smoke | Black |
| 86 | 0 | 33 | 155 | 3 | 0 | 0 | 0 | 0 | 3 | 2551 | No Smoke | Other |
| 87 | 0 | 20 | 105 | 1 | 1 | 0 | 0 | 0 | 1 | 2557 | Smoke | White |
| 88 | 0 | 21 | 108 | 1 | 1 | 0 | 0 | 1 | 2 | 2594 | Smoke | White |
| 89 | 0 | 18 | 107 | 1 | 1 | 0 | 0 | 1 | 0 | 2600 | Smoke | White |
| 91 | 0 | 21 | 124 | 3 | 0 | 0 | 0 | 0 | 0 | 2622 | No Smoke | Other |
To plot a barchart, we are going to make use of geom_bar(), but first we would need to set our global aes() first! In our case, we would want our x-axis to be raceF, y-axis to be birth weight in grams (bwt) and to group the barcharts together we can assign smokeR to fill.
Editor note: the
statingeom_bar()can either be "identity" or "summary". "identity" is used if the numbers used in the dataframe are the numbers you want displayed on the graph. In our example, we need to use "summary" as we would like to plot the mean birth weight!positionargument is the same asgeom_hist().widthargument just changes the width of the bar graph.
birthwt_bar = ggplot(birthwt, aes(x = raceF, y=bwt, fill=smokeR)) +
geom_bar(stat="summary",position="dodge", width = 0.5, fun = mean)+
#Beautifying the graph#
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Race') + ylab('Baby Birth Weight (g)') +
labs(title = "Baby Birth Weight (g) vs Race", subtitle = 'grouped by Smoking Status') +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20))+
theme(strip.text = element_text(size = 20))+
theme(legend.text = element_text(size = 20)) +
theme(legend.title = element_text(size = 25))+ labs(fill='Smoking Status') +
theme(plot.subtitle = element_text(size=20))+
theme(legend.position = c(0.9, 0.9))+
theme(legend.background=element_blank())
birthwt_bar
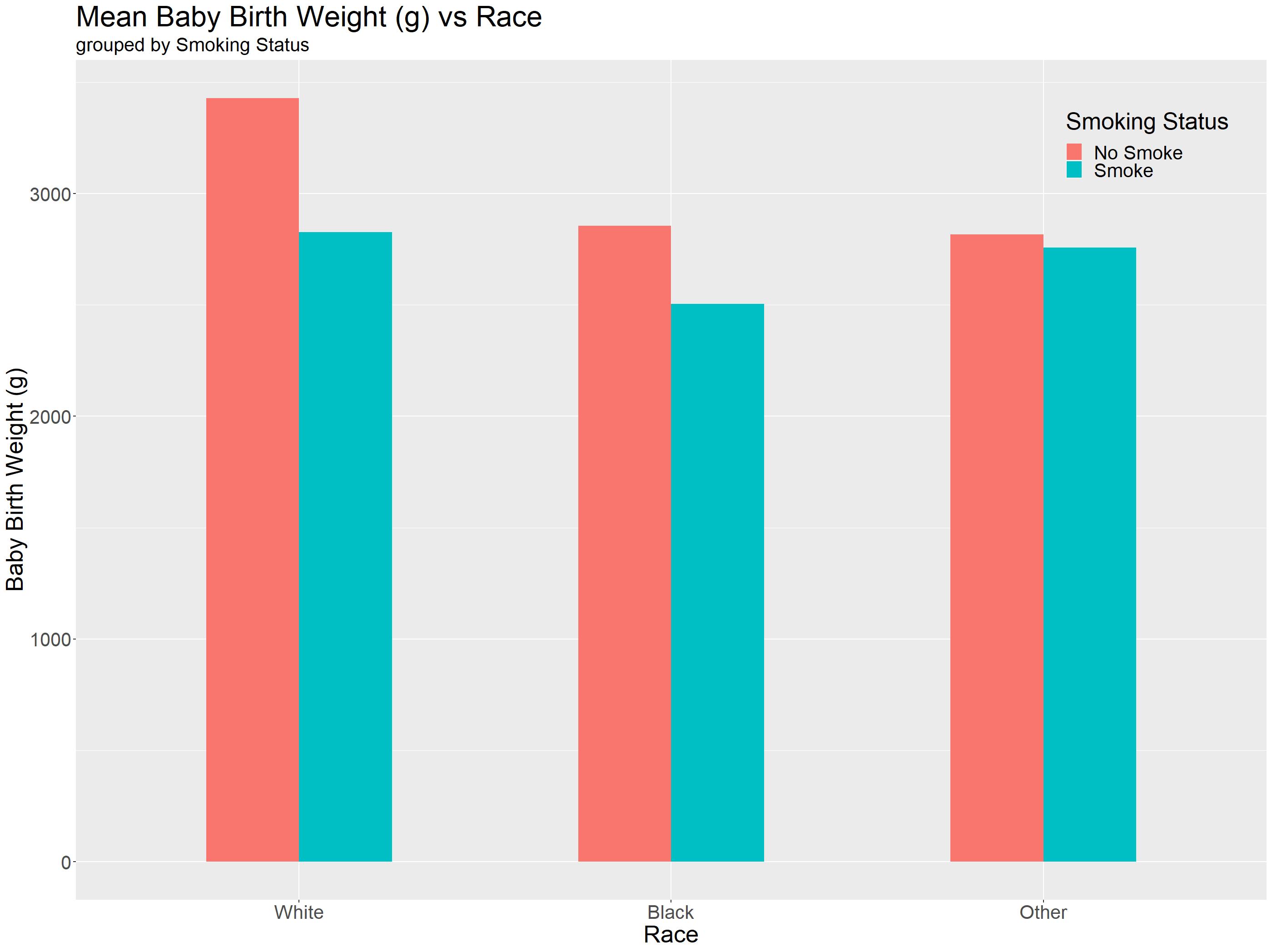
We are almost done! With a quick glance, anyone can immediately understand the following points:
- Non-smoking mothers will have the highest baby weight across all races
- Mothers whose race is ‘White' will produce the heaviest baby regardless of their smoking status
What we are missing now are some errorbars! Luckily for us this website provided us with a handy code snippet to help us summarise our data nicely!
What we can plot for errorbars are Confidence Intervals (CI), Standard Deviation (sd) or standard error (se). In our tutorial, we are going to define our own standard error functionstd() and use it to plot our errorbars!
std <- function(x) sd(x)/sqrt(length(x))
data_summary <- function(data, varname, groupnames){
require(plyr)
summary_func <- function(x, col){
c(mean = mean(x[[col]], na.rm=TRUE), se = std(x[[col]]))
}
data_sum<-ddply(data, groupnames, .fun=summary_func, varname)
data_sum <- rename(data_sum, c("mean" = varname))
return(data_sum)
}
#it takes in the dataframe, varname takes in y-axis, groupnames take in a vector of grouping we want, in our case race and smoking status!
df2 <- data_summary(birthwt, varname="bwt", groupnames=c("raceF", "smokeR"))
df2
| se | bwt | smokeR | raceF | |
|---|---|---|---|---|
| 1 | 107.05144 | 3428.750 | No Smoke | White |
| 2 | 86.87610 | 2826.846 | Smoke | White |
| 3 | 155.31358 | 2854.500 | No Smoke | Black |
| 4 | 201.45504 | 2504.000 | Smoke | Black |
| 5 | 95.64864 | 2815.782 | No Smoke | Other |
| 6 | 233.83975 | 2757.167 | Smoke | Other |
Now we can plot, the thing to take note here is the use of stat='identity' as our mean is precalculated!
birthwt_bareb = ggplot(df2 , aes(x = raceF, y=bwt, fill=smokeR)) +
geom_bar(stat="identity", position='dodge', width = 0.5)+
geom_errorbar(aes(ymin=bwt-se, ymax=bwt+se), width=.2, position=position_dodge(.5))+
#Standard graph aesthetics
theme() +
theme(axis.text=element_text(size=20)) +
theme(axis.title=element_text(size=25)) +
xlab('Race') + ylab('Baby Birth Weight (g)') +
labs(title = "Mean Baby Birth Weight (g) vs Race", subtitle = 'grouped by Smoking Status') +
theme(plot.title = element_text(size=30)) +
theme(plot.subtitle = element_text(size=20))+
theme(strip.text = element_text(size = 20))+
theme(legend.text = element_text(size = 20)) +
theme(legend.title = element_text(size = 25))+ labs(fill='Smoking Status') +
theme(plot.subtitle = element_text(size=20))+
theme(legend.position = c(0.9, 0.9))+
theme(legend.background=element_blank())
birthwt_bareb
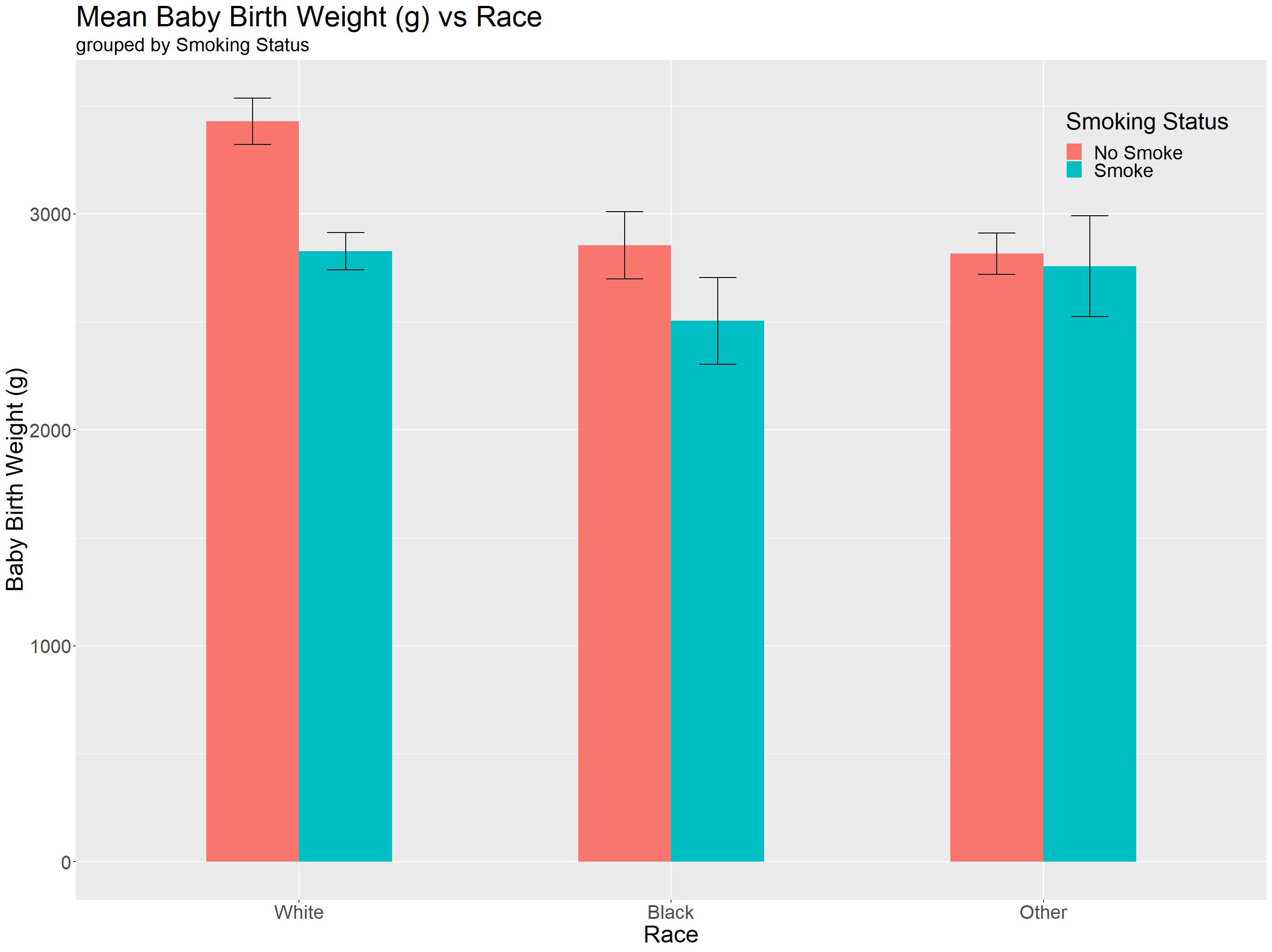
Editor note: Even though you could observe such things in a graph, proper statistical testing needs to be done to conclude if the differences are significant!