Exploration of COVID19 dataset: Time-Series daily cases plot
Recap & Grabbing Time-Series Data
First, customary importing of packages and data if you haven't already done so from the previous sections.
import pandas as pd
import matplotlib.pyplot as plt
ASEAN_countries_list = ['Brunei', 'Cambodia', 'Indonesia', 'Laos', 'Malaysia', 'Burma', 'Philippines', 'Singapore', 'Vietnam']
link = 'https://raw.githubusercontent.com/CSSEGISandData/COVID-19/master/csse_covid_19_data/csse_covid_19_time_series/time_series_covid19_confirmed_global.csv'
covid19_data = pd.read_csv(link)
asean_df = covid19_data[covid19_data['Country/Region'].isin(ASEAN_countries_list)]
asean_df.set_index('Country/Region', inplace = True)
asean_df_dropped = asean_df.drop(columns = ['Province/State', 'Lat', 'Long'])
asean_t = asean_df_dropped.T
asean_t
| Country/Region | Brunei | Burma | Cambodia | Indonesia | Laos | Malaysia | Philippines | Singapore | Vietnam |
|---|---|---|---|---|---|---|---|---|---|
| 1/22/20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1/23/20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| 1/24/20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 |
| 1/25/20 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 3 | 2 |
| 1/26/20 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 4 | 2 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7/30/21 | 336 | 294460 | 76585 | 3372374 | 5919 | 1095486 | 1580824 | 64861 | 141122 |
| 7/31/21 | 337 | 299185 | 77243 | 3409658 | 6299 | 1113272 | 1588965 | 64981 | 150060 |
| 8/1/21 | 337 | 302665 | 77914 | 3440396 | 6566 | 1130422 | 1597689 | 65102 | 157507 |
| 8/2/21 | 338 | 306354 | 78474 | 3462800 | 6765 | 1146186 | 1605762 | 65213 | 157507 |
| 8/3/21 | 338 | 311067 | 79051 | 3496700 | 7015 | 1163291 | 1612541 | 65315 | 174461 |
560 rows × 9 columns
Using daily cumulative cases to calculate daily increase
Previously we looked at simple plotting of cumulative cases against dates. What if we would want to look at daily cases instead? We would need to do some maths and manipulation (i promise its nothing too scary).
Imagine our dataframe for 1 country now looks like this:
| Day | Cases |
|---|---|
| 0 | 0 |
| 1 | 0 |
| 2 | 1 |
| 3 | 3 |
| 4 | 3 |
| 5 | 4 |
How we are going to do this is to make make a new dataframe which brings a row to align with the previous day's data. So we can do some quick maths later!
| Day | Cases_O | Cases_ShiftF |
|---|---|---|
| 1 | 0 | NaN |
| 2 | 1 | 0 |
| 3 | 3 | 1 |
| 4 | 3 | 3 |
| 5 | 4 | 3 |
Then by adding a new column called "difference" we can calculate the difference / increase in cases for each day!
| Day | Cases_O | Cases_ShiftF | difference |
|---|---|---|---|
| 1 | 0 | NaN | NaN |
| 2 | 1 | 0 | 1 |
| 3 | 3 | 1 | 2 |
| 4 | 3 | 3 | 0 |
| 5 | 4 | 3 | 1 |
We can then plot the difference!
We can try do this for Singapore first.
import numpy as np
#Make a copy so we don't actually change the previous dataframe
sg_s = asean_t['Singapore'].copy()
#Use this to shift the entire column down by 1
sg_s_F = sg_s.shift(1)
#Merge the two columns by using the same dates
sg_df = pd.concat([sg_s, sg_s_F], axis=1)
#Rename the columns for easier reference later
sg_df.columns = ['Cases_O', 'Cases_ShiftF']
#Creating a new column
sg_df['difference'] = sg_df['Cases_O'] - sg_df['Cases_ShiftF']
sg_df
| Cases_O | Cases_ShiftF | difference | |
|---|---|---|---|
| 1/22/20 | 0 | NaN | NaN |
| 1/23/20 | 1 | 0.0 | 1.0 |
| 1/24/20 | 3 | 1.0 | 2.0 |
| 1/25/20 | 3 | 3.0 | 0.0 |
| 1/26/20 | 4 | 3.0 | 1.0 |
| ... | ... | ... | ... |
| 7/30/21 | 64861 | 64722.0 | 139.0 |
| 7/31/21 | 64981 | 64861.0 | 120.0 |
| 8/1/21 | 65102 | 64981.0 | 121.0 |
| 8/2/21 | 65213 | 65102.0 | 111.0 |
| 8/3/21 | 65315 | 65213.0 | 102.0 |
560 rows × 3 columns
If we just refer to the code snippet above, we can easily make a function that does this for each country! Lets call this function country_diff() which takes in the original dataframe (asean_t) and the country of choice. In turn, make it return a new modified dataframe!
def country_diff(df, country):
#Make a copy so we don't actually change the previous dataframe
df_s = df[country].copy()
#Use this to shift the entire column down by 1
df_s_F = df_s.shift(1)
#Merge the two columns by using the same dates
country_df = pd.concat([df_s, df_s_F], axis=1)
#Rename the columns for easier reference later
country_df.columns = [f'{country}_Cases_O', f'{country}_Cases_ShiftF']
#Creating a new column
country_df[f'{country}_difference'] = country_df[f'{country}_Cases_O'] - country_df[f'{country}_Cases_ShiftF']
return country_df
#Test the function
#brunei_df = country_diff(asean_t, 'Brunei')
Notice that our function is generally the same as our Singapore example, except we made some changes in the naming conventions (you'll see why later) to better reference them later!
We can now make use of ASEAN_countries_list to grab the daily cases of all our countries!
We can then make use of the pd.concat function again to have a dataframe consisting solely of daily cases for each country!
#Create a new dataframe
asean_daily_df = pd.DataFrame()
#Run a for loop across all countries
#Reminder:
#ASEAN_countries_list = ['Brunei', 'Cambodia', 'Indonesia', 'Laos',
# 'Malaysia', 'Burma', 'Philippines', 'Singapore', 'Vietnam']
for country in ASEAN_countries_list:
#Run the function and storing it into a temporary dataframe
#per country
temp_df = country_diff(asean_t, country)
#Attach this temporary df to the actual dataframe
#While selecting for the "Difference" column only!
asean_daily_df = pd.concat([asean_daily_df, temp_df[f'{country}_difference']],sort = False, axis = 1)
#Drop the very first row because it makes no sense to have a change between 0th day and -1th day
asean_daily_df = asean_daily_df.iloc[1:,]
asean_daily_df
| Brunei_difference | Cambodia_difference | Indonesia_difference | Laos_difference | Malaysia_difference | Burma_difference | Philippines_difference | Singapore_difference | Vietnam_difference | |
|---|---|---|---|---|---|---|---|---|---|
| 1/23/20 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 |
| 1/24/20 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 1/25/20 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1/26/20 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 1/27/20 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7/30/21 | 3.0 | 668.0 | 41168.0 | 244.0 | 16840.0 | 5127.0 | 8537.0 | 139.0 | 7717.0 |
| 7/31/21 | 1.0 | 658.0 | 37284.0 | 380.0 | 17786.0 | 4725.0 | 8141.0 | 120.0 | 8938.0 |
| 8/1/21 | 0.0 | 671.0 | 30738.0 | 267.0 | 17150.0 | 3480.0 | 8724.0 | 121.0 | 7447.0 |
| 8/2/21 | 1.0 | 560.0 | 22404.0 | 199.0 | 15764.0 | 3689.0 | 8073.0 | 111.0 | 0.0 |
| 8/3/21 | 0.0 | 577.0 | 33900.0 | 250.0 | 17105.0 | 4713.0 | 6779.0 | 102.0 | 16954.0 |
559 rows × 9 columns
Edit on 15th August 2021, I found out that there is a function called
pd.diff()which does this exact thing. I will now demonstrate it
# Yes thats it...
asean_daily_diff = asean_t.diff().iloc[1:,]
asean_daily_diff
| Brunei_difference | Cambodia_difference | Indonesia_difference | Laos_difference | Malaysia_difference | Burma_difference | Philippines_difference | Singapore_difference | Vietnam_difference | |
|---|---|---|---|---|---|---|---|---|---|
| 1/23/20 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 2.0 |
| 1/24/20 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.0 | 0.0 |
| 1/25/20 | 0.0 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 1/26/20 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| 1/27/20 | 0.0 | 1.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | 0.0 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 7/30/21 | 3.0 | 668.0 | 41168.0 | 244.0 | 16840.0 | 5127.0 | 8537.0 | 139.0 | 7717.0 |
| 7/31/21 | 1.0 | 658.0 | 37284.0 | 380.0 | 17786.0 | 4725.0 | 8141.0 | 120.0 | 8938.0 |
| 8/1/21 | 0.0 | 671.0 | 30738.0 | 267.0 | 17150.0 | 3480.0 | 8724.0 | 121.0 | 7447.0 |
| 8/2/21 | 1.0 | 560.0 | 22404.0 | 199.0 | 15764.0 | 3689.0 | 8073.0 | 111.0 | 0.0 |
| 8/3/21 | 0.0 | 577.0 | 33900.0 | 250.0 | 17105.0 | 4713.0 | 6779.0 | 102.0 | 16954.0 |
559 rows × 9 columns
Plotting Daily Cases for each country
With this new dataframe, we can now make use of the plotting function in the previous section for time-series!
Note the use of f'' to easily specify the different columns :)
fig, ax = plt.subplots(figsize = (10,10))
#Iterate through our countries so we can plot automatically plot them!
for country in ASEAN_countries_list:
asean_daily_df[f'{country}_difference'].plot(ax=ax, label = country)
ax.set_ylabel('Confirmed Daily Cases')
ax.set_xlabel('Dates')
ax.legend()
plt.show()
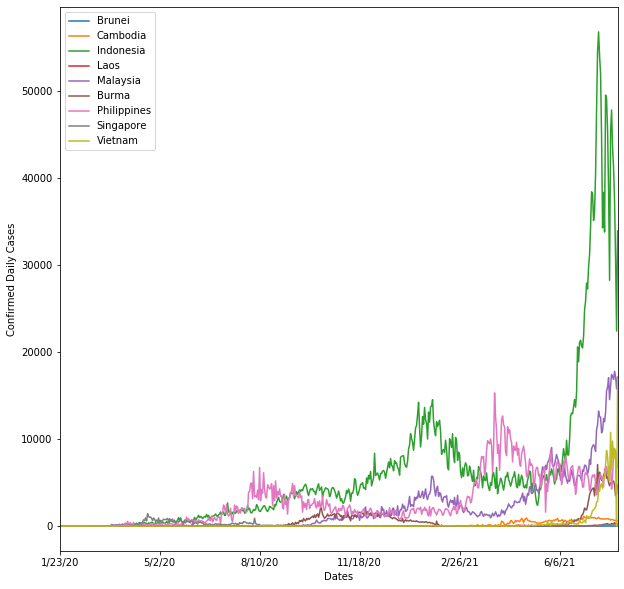
From the graph above, you can tell that recently, Indonesia has the highest confirmed daily cases. Followed by Malaysia and Vietnam.
The interesting thing is that over time, we can tell that Indonesia's daily cases are slowly decreasing.